Why is open sharing of epigenomic (or genomic) data so important for the community?
Without it, many millions of hours and dollars spent on research would be lost. Researchers would have to produce new data which would otherwise already be available, slowing down the overall progress of science.
It is also efficient in that it allows scrutiny by scientists who are not directly involved in the research and potential repurposing of data to address other research questions. It might also encourage more researchers to get involved in the big science of genomics/epigenomics.
What are some of the issues involved in this type of open data?
The main issues have to do with scientific competition and the protection of research participants who have contributed their data to research. Our study focuses on the latter: how might sharing data in different ways affect the privacy and welfare of research participants?
Epigenomic data raises serious questions about anti-discrimination protections and informed consent. As most often there are risks and benefits to research participants with any course of action, the challenge is to carefully assess and balance these.
What does your study show?
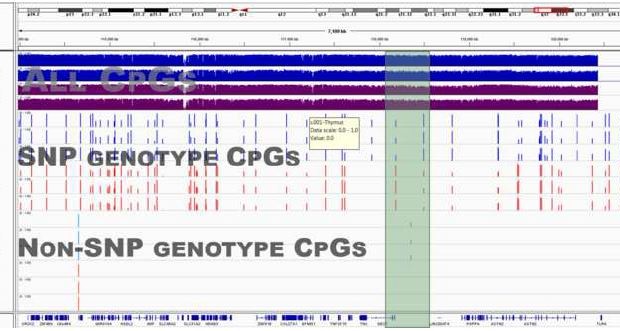
Our study shows that a lot of epigenomic data can be shared publicly. We quantified the amount of non-CpG DNA sequence information that can be inferred from the most detailed methylation data currently being produced.
Our study shows that a lot of epigenomic data can be shared publicly.
The results show that most of it can reasonably be removed from open-access datasets to reduce the risk of re-identification of research data from anonymous participants.
We also assessed the harms that might occur as a result of data re-identification, notably through loss of privacy regarding health and demographic data that is provided with samples. We therefore offer guidance for open-access releases that would reduce potential harmful consequences too.
What guidelines are currently being used for this type of data, and what new guidelines do you propose?
To be honest, there aren’t any established guidelines in this area. Epigenomic data has not yet been produced on a scale comparable to that being generated by the International Human Epigenome Consortium of which we are members. That is why we decided to do this research.
Based on our study – which we hope might inform the development of community guidelines – we recommend filtering known direct single-nucleotide polymorphism influences on methylation data, along with some demographic and rare disease information that presents greater risk. This data should still be shared but with additional protections.
- What can we learn from single cells? - 1st June 2016
- Scaling down to single cells - 14th December 2015
- Guidance for the open sharing of epigenome data: an author Q+A - 22nd July 2015
Comments