
Keiko Herrick and colleagues at the University of Alaska Fairbanks have put forward a model of avian influenza virus (AIV) prediction in wild birds, published yesterday in Veterinary Research.
This is the first large-scale large ecological niche model for avian influenza in wild birds based on machine-learning algorithms. The authors mined surveillance data for 2005-2010 from the Influenza Research Database, which yielded a large set of georeferenced sample points, complete with AIV detection status, viral subtype and other related parameters.
The sampling data was then layered into ArcMap, along with 41 predictor layers – accounting for environmental and anthropogenic variables such as geographic elevation, adjusted mean temperatures and human population density – all obtained from open source projects.
In the next step, the data and variable layers were classified using the Random Forests machine-learning algorithm, resulting in a global ecological model of avian influenza prediction in wild birds. The model also predicted an AIV-positive niche in regions with low annual rainfall and low temperatures, primarily in northern regions with a continental climate such as Siberia and interior Alaska.
The model of avian influenza prediction reached by the authors, while not definitive, is a first accurate and global-scale predictive model. Additional statistical analysis, including a Receiver Operating Characteristic curve, indicated that the model was 79% accurate for training data, 76% for test data.
This is one of the few studies in the field to consider various wild bird populations and all available sub-types of avian influenza. Although public attention has been drawn to H5N1 outbreaks in poultry – linked to cases of infection in humans – the virus is considerably more prevalent in wild bird populations, to the extent that they form the main genetic reservoir of avian influenza.
Keiko Herrick describes the background of the study in more detail:
“The recent outbreaks of H7N9 in humans underscore the importance of including subtypes other than highly pathogenic H5N1 in avian influenza research. A lot of excellent models of H5N1 transmission and risk factors have been constructed, so it was time to start looking at AIV in wild bird populations on a large scale.
Ensemble data-mining is a powerful way to define the ecological niche of AIV in wild birds. In addition, machine-learning methods are a new way to study the vast amounts of surveillance data that are being collected.
IRD is a very valuable resource for this kind of research because it combines spatial data in addition to genetic and disease data. This study is a preliminary model that correlates AIV-positivity with bioclimatic variables, so ideally, it would guide future surveillance and data collection, which would provide ground-truth for the model, strengthen the prediction, and begin to address the mechanisms underlying the correlation.”
The following map, presented in Robinson (sphere) projection, central meridian 145°, is a global representation of the predicted Relative Occurrence Index (ROI) of avian influenza virus in wild birds.
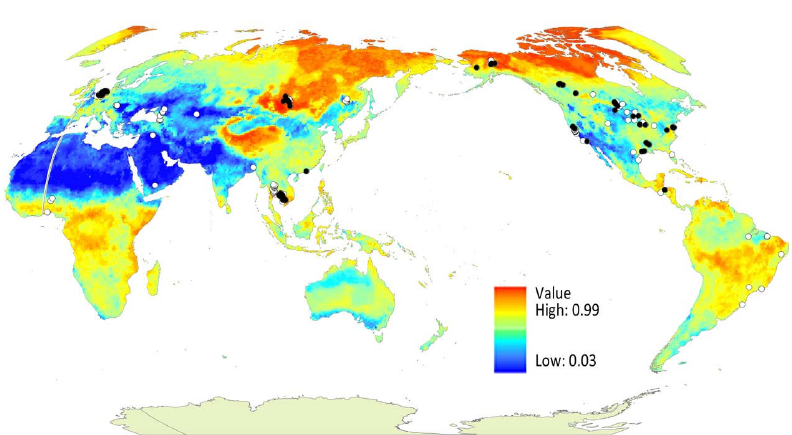
The map and underlying ROI data suggest that many northern regions may have the highest risk of avian influenza outbreak, relative to other locations. The model could guide future AIV surveillance and sampling efforts. For instance Siberia and Northern Canada were identified as areas of high relative risk in the model, but have hitherto had minimal surveillance for signs of AIV.
This research presents a novel proof-of-concept and was made available to all readers in an open access journal, in line with the authors’ objective to encourage greater cooperation and data sharing in the avian influenza virus research community.
Future collaborative sampling efforts will strengthen the model, its predictive value and our collective understanding of avian influenza virus ecology.
The full article is available at https://www.veterinaryresearch.org/content/44/1/42/.
Matt Landau
Journal Development Editor, Veterinary Research
@Matt_Landau
For more information about BioMed Central’s Animal and Plant Science journals, please follow us on Twitter.
Matthew Landau
Latest posts by Matthew Landau (see all)
- March biology highlights: octopuses, heavy metal, and pocket-sized DNA sequencing - 10th April 2015
- From farm to plate, make food safe! - 7th April 2015
- Dried up! The effects of 2014 on Western US Lake Ecosystems - 13th November 2014
Comments