Genomics charts new territory today – your nearest pond – with the publication of the water-flea (Daphnia pulex) genome sequence in Science. Intriguingly, the diminutive water-flea contains at least 31,000 genes – more than have so far been found in any other animal – and the genome release marks the first crustacean genome to be completed. 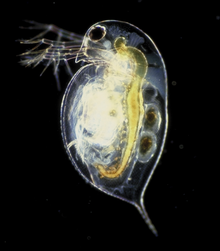 Daphnia’s high gene number is thought to arise from rapid duplication of genetic material. Project leader, John Colbourne said: "We estimate a rate that is three times greater than those of other invertebrates and 30 percent greater than that of humans."
Daphnia’s high gene number is thought to arise from rapid duplication of genetic material. Project leader, John Colbourne said: "We estimate a rate that is three times greater than those of other invertebrates and 30 percent greater than that of humans."
As the water-flea is already a model organism for ecology, toxicology and evolutionary biology, the Daphnia genome allows these disciplines to move forward, with detailed studies of gene-environment interactions (environmental genomics) in particular. More than thirty research articles have been authored during the project collaboration, in a companion series to the Daphnia genome. Each related article is published by BioMed Central in an open access journal and is freely available online. A complete publication list can be found on the series homepage and a Commentary by Diethard Tautz is also published today in BMC Biology.
The genome data has made possible experiments that demonstrate the species’ renewed importance as a sentinel of freshwater ecosystem health. For example, experiments have explored the genetic response of Daphnia to environmental stressors such as temperature acidity and potential environmental contaminants, including heavy metals and pharmaceutical agents.
The Daphnia genome also allows us to investigate the evolution of peculiarities, such as its clonal reproductive mode. As a representative of the crustacea, genetic comparison with other groups provides further insight into invertebrate taxonomy and the evolutionary development of novel features, including pancrustacean eyes and segmented body plans. Since Daphnia behavior is so well-studied, the results of transcriptional profiling can now be used to measure the genetic and phenotypic response of these organisms to predators.
Perhaps the most fascinating aspect of the Daphnia genome is its flexibility from an evolutionary perspective. The genome is compact with relatively short gaps (introns) between the gene sections that code for proteins. The unprecedented gene count and duplication rate could be a byproduct of recent reproductive events in the pedigree of the single individual (dubbed "The Chosen One") that was selected from a naturally inbred population for sequencing. Another appealing hypothesis suggested by the investigators is based on the observation that Daphnia’s duplicated genes can gain a new function remarkably quickly: the acquisition of a useful stream of fresh genetic material could be an evolutionary answer to the challenges posed by the fluctuating freshwater conditions in which Daphnia populations survive.
John K. Colbourne1,*, Michael E. Pfrender2,†, Donald Gilbert1,3, W. Kelley Thomas4, Abraham Tucker3,4, Todd H. Oakley5, Shinichi Tokishita6, Andrea Aerts7, Georg J. Arnold8, Malay Kumar Basu9,‡, Darren J. Bauer4, Carla E. Cáceres10, Liran Carmel9,§ (2011). The Ecoresponsive Genome of Daphnia pulex Science, 331, 555-561 : 10.1126/science.1197761
Photo: Paul Hebert
- Biotechnology for a bio-based economy - 5th June 2019
- Sustainable energy at the American Chemical Society Spring meeting - 11th April 2018
- Biotechnology for Biofuels – Special Issue on Life Cycle Analysis - 17th May 2017
Comments