This week a few of us here at BioMed Central are off to Boston for ISCB’s annual ISMB conference and its Sp ecial Interest Group (SIG) meetings. First thing’s first: We’ll be giving away cool swag like our new Bruce Lee/Kill Bill inspired GigaScience Open Data tshirts. Our 8GB GigaPanda USB drives will also be making a repeat appearance.
ecial Interest Group (SIG) meetings. First thing’s first: We’ll be giving away cool swag like our new Bruce Lee/Kill Bill inspired GigaScience Open Data tshirts. Our 8GB GigaPanda USB drives will also be making a repeat appearance.
Where can you get these? We’ll be at booths #419 and #420 and would love to chat to you about open peer-review and our journals Biology Direct and GigaScience or some of our informatics journals like BMC Bioinformatics, Journal of Biomedical Semantics, and BioData Mining. GigaScience will be celebrating their second birthday at the meeting, and our Editors Laurie Goodman and Scott Edmunds will be around to talk to you about their innovative work publishing data and workflows, our call for Technical Notes and our new theme series on Optical Mapping with David Schwartz.
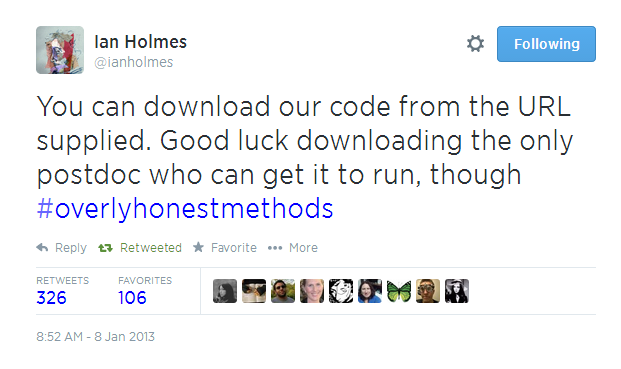
But it’s not just about the cool swag. ISMB is also the chance for us to engage with the way open science is changing research and the way that research is communicated. Like last year, we’re organising a workshop with ResearchObject, Nanopub.org, and ISA-Tab on What Bioinformaticians Need to Know Beyond the PDF (inspired by Force 11’s Beyond the PDF). We’ll be joined by our usual overqualified list of speakers—Phil Bourne, Kaitlin Thaney, Francis Ouellette, and others—to talk about reproducibility, open peer review, credit for source code, and data publishing. Ciaran O’Neill and myself will be talking about new models of peer review and the opportunities and challenges of peer review of software. The credit for and peer-review of software and source code is particularly of interest to us and something we’ve recently spoken at length about with Carole Goble at our recent Editor’s Day Conference. In the video below, for example, Carole addresses the well-known though slightly embarrassing issue of having access to the software or source code versus actually being able to get it to run.

Quoting Ian Holmes, she says “ You can download our code from the URL supplied. Good luck downloading the only postdoc who can get it to run.”
An answer to this challenge might be a more inclusive and tiered system of peer review, bringing into the fold the PhD and postdocs actually able to run the software.
We’ll be soliciting discussion from the audience on this point and others, as well as handing out a few freebies there as well, so join us on Tuesday at 2pm in Ballroom C to join in on the discussion.
Special Interest Group Meetings
ISMB is well known for its SIGs alone. Of particular interest to us here at BioMed Central are BOSC (the Bioinformatics Open Source Conference), the AFP (Automated Function Prediction), and the Bio-Ontologies SIGs.
BOSC
A sponsor of BOSC, with it
s strong Open Source focus GigaScience aims to be a formal home for the kind of discussions happening at BOSC. Topics to be discussed this year include Software Interoperability, Visualisation, Genome-scale Data and Beyond, and Open Science and Reproducible Research. We’re looking forward
to this year’s keynotes:
- “A History of Bioinformatics (in the year 2039)” by Titus Brown
- “Biomedical Research as an Open Digital Enterprise” by Phil Bourne,
Make sure to also check out talks by Karthik Ram (“Fostering the next generation of data-driven open science with R”) and Ross Mounce (“PLUTo: Phyloinformatic Literature Unlocking Tools”). Like in previous years, this year’s BOSC will be preceded by a two-day Codefest (this Wednesday and Thursday at hack/reduce). It’s free and all are welcome to attend.
Bio-Ontologies
One of our workshop co-organisers and long-time ISA-Tab advisor to GigaScience, Philippe Rocca-Sera, is also co-organising the Bio-Ontologies SIG. This long-running SIG aims to provide insight into the latest research in ontologies and more generally the organisation, presentation and dissemination of knowledge in biomedicine and the life sciences. Our Journal of Biomedical Semantics has become a home for such topics in our recent thematic series on Bio-ontologies, which includes talks from last year’s Bio-Ontologies SIG. Also check out GigaScience’s previous blog on the SIG.
AFP
An increasing concern with the recent deluge in genomic sequencing data is the inherent difficulty and expense that exists to scale-up and biologically characterise the plethora of genes and proteins of unknown function – one problem of which is computational annotation of protein function. The mission of the AFP SIG is to bring together computational biologists, experimental biologists, and biocurators dealing with gene and gene product function prediction, to share ideas and create collaborations. This year they are also conducting a new multi-year Critical Assessment of Functional Annotation, or CAFA, experiment. Keynotes for the AFP SIG will be given by Philip Bourne, Fiona Brinkman, and Mark Gerstein. Papers from AFP talks are being published in a GigaScience series, where a few papers from last year’s AFP currently live.
Having a long history as a particularly social media savvy conference, we’ll all be on Twitter and our various handles are below. Follow the action from the hashtags #BOSC2014 and #ISMB2014, and feel free to tweet us if you’d like to talk about any of the open science work we’ve been up to at BioMed Central. See you in Boston!
Scott Edmunds, GigaScience: @GigaScience
Clare Garvey, Genome Biology: @GenomeBiology
Laurie Goodman, GigaScience: @GigaScience
Amye Kenall, Open Data BMC: @AmyeKenall and @OpenDataBMC
Ciaran O’Neill, Biology Direct and Biome: @cjmoneill
Comments